Rows: 1,470
Columns: 31
$ Age <int> 41, 49, 37, 33, 27, 32, 59, 30, 38, 36, 35, 2…
$ Attrition <fct> Yes, No, Yes, No, No, No, No, No, No, No, No,…
$ BusinessTravel <fct> Travel_Rarely, Travel_Frequently, Travel_Rare…
$ DailyRate <int> 1102, 279, 1373, 1392, 591, 1005, 1324, 1358,…
$ Department <fct> Sales, Research_Development, Research_Develop…
$ DistanceFromHome <int> 1, 8, 2, 3, 2, 2, 3, 24, 23, 27, 16, 15, 26, …
$ Education <ord> College, Below_College, College, Master, Belo…
$ EducationField <fct> Life_Sciences, Life_Sciences, Other, Life_Sci…
$ EnvironmentSatisfaction <ord> Medium, High, Very_High, Very_High, Low, Very…
$ Gender <fct> Female, Male, Male, Female, Male, Male, Femal…
$ HourlyRate <int> 94, 61, 92, 56, 40, 79, 81, 67, 44, 94, 84, 4…
$ JobInvolvement <ord> High, Medium, Medium, High, High, High, Very_…
$ JobLevel <int> 2, 2, 1, 1, 1, 1, 1, 1, 3, 2, 1, 2, 1, 1, 1, …
$ JobRole <fct> Sales_Executive, Research_Scientist, Laborato…
$ JobSatisfaction <ord> Very_High, Medium, High, High, Medium, Very_H…
$ MaritalStatus <fct> Single, Married, Single, Married, Married, Si…
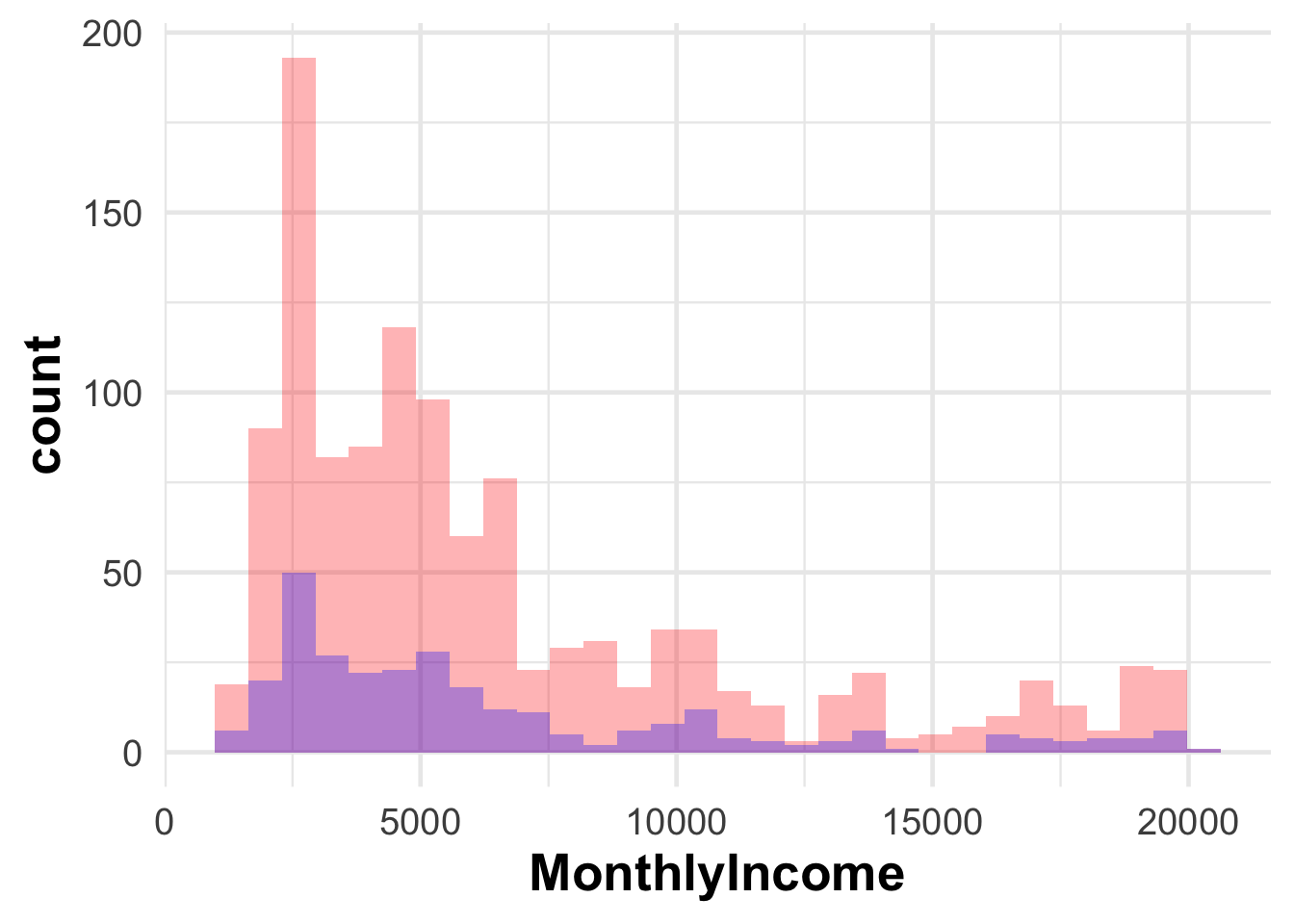
$ MonthlyIncome <int> 5993, 5130, 2090, 2909, 3468, 3068, 2670, 269…
$ MonthlyRate <int> 19479, 24907, 2396, 23159, 16632, 11864, 9964…
$ NumCompaniesWorked <int> 8, 1, 6, 1, 9, 0, 4, 1, 0, 6, 0, 0, 1, 0, 5, …
$ OverTime <fct> Yes, No, Yes, Yes, No, No, Yes, No, No, No, N…
$ PercentSalaryHike <int> 11, 23, 15, 11, 12, 13, 20, 22, 21, 13, 13, 1…
$ PerformanceRating <ord> Excellent, Outstanding, Excellent, Excellent,…
$ RelationshipSatisfaction <ord> Low, Very_High, Medium, High, Very_High, High…
$ StockOptionLevel <int> 0, 1, 0, 0, 1, 0, 3, 1, 0, 2, 1, 0, 1, 1, 0, …
$ TotalWorkingYears <int> 8, 10, 7, 8, 6, 8, 12, 1, 10, 17, 6, 10, 5, 3…
$ TrainingTimesLastYear <int> 0, 3, 3, 3, 3, 2, 3, 2, 2, 3, 5, 3, 1, 2, 4, …
$ WorkLifeBalance <ord> Bad, Better, Better, Better, Better, Good, Go…
$ YearsAtCompany <int> 6, 10, 0, 8, 2, 7, 1, 1, 9, 7, 5, 9, 5, 2, 4,…
$ YearsInCurrentRole <int> 4, 7, 0, 7, 2, 7, 0, 0, 7, 7, 4, 5, 2, 2, 2, …
$ YearsSinceLastPromotion <int> 0, 1, 0, 3, 2, 3, 0, 0, 1, 7, 0, 0, 4, 1, 0, …
$ YearsWithCurrManager <int> 5, 7, 0, 0, 2, 6, 0, 0, 8, 7, 3, 8, 3, 2, 3, …