infant_mortality_df <- readxl::read_excel("../data/demo_minfind.xls")A whole game
Readings and class materials for Tuesday, September 19, 2023
You can access an Excel spreadsheet on infant mortality downloaded from Eurostat. We discovered last week that utilizing the Eurostat data is significantly facilitated by the eurostat package; however, for the purpose of practical demonstration, we now need to showcase the multitude of cleaning procedures required to establish links between series originating from diverse sources.
Lets download the infant mortality rates from Eurostat (in Excel format) and link it with the fertility_df
The issue here is that there are multiple tables on the first sheet (not a rare thing).
infant_mortality_df# A tibble: 339 × 62
Infant mortality rate…¹ ...2 ...3 ...4 ...5 ...6 ...7 ...8 ...9 ...10
<chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
2 Last update 4474… <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
3 Extracted on 4482… <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
4 Source of data Euro… <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
5 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
6 INDIC_DE Earl… <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
7 UNIT Rate <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
8 <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA> <NA>
9 GEO/TIME 1960 1961 1962 1963 1964 1965 1966 1967 1968
10 European Union - 27 co… : : : : : : : : :
# ℹ 329 more rows
# ℹ abbreviated name: ¹`Infant mortality rates [demo_minfind]`
# ℹ 52 more variables: ...11 <chr>, ...12 <chr>, ...13 <chr>, ...14 <chr>,
# ...15 <chr>, ...16 <chr>, ...17 <chr>, ...18 <chr>, ...19 <chr>,
# ...20 <chr>, ...21 <chr>, ...22 <chr>, ...23 <chr>, ...24 <chr>,
# ...25 <chr>, ...26 <chr>, ...27 <chr>, ...28 <chr>, ...29 <chr>,
# ...30 <chr>, ...31 <chr>, ...32 <chr>, ...33 <chr>, ...34 <chr>, …After exploring the data, you may realize that the name of the data is always in the second column. Our table starts after where find the “Infant mortality rate”.
pull(infant_mortality_df, 2) # 2nd column as vector [1] NA "44742.521828703699"
[3] "44822.86385927083" "Eurostat"
[5] NA "Early neonatal mortality rate"
[7] "Rate" NA
[9] "1960" ":"
[11] ":" ":"
[13] ":" ":"
[15] "17.100000000000001" "10.9"
[17] "10.699999999999999" "13.9"
[19] "20.800000000000001" "19.699999999999999"
[21] "9.5999999999999996" "16.100000000000001"
[23] "12.300000000000001" "15.9"
[25] ":" "14.6"
[27] "21" "17.800000000000001"
[29] ":" ":"
[31] "7.2999999999999998" "16.300000000000001"
[33] "22.100000000000001" ":"
[35] "11.9" "20.199999999999999"
[37] ":" "15"
[39] ":" "15.6"
[41] "10.300000000000001" "12.6"
[43] "11.800000000000001" ":"
[45] ":" ":"
[47] ":" "7.0999999999999996"
[49] ":" "9.9000000000000004"
[51] "14.4" "13.699999999999999"
[53] ":" "18.600000000000001"
[55] ":" ":"
[57] ":" ":"
[59] ":" "17.300000000000001"
[61] ":" ":"
[63] ":" ":"
[65] ":" ":"
[67] ":" ":"
[69] NA NA
[71] "not available" NA
[73] "Infant mortality rate" "Rate"
[75] NA "1960"
[77] ":" ":"
[79] ":" ":"
[81] ":" "31.399999999999999"
[83] "45.100000000000001" "20"
[85] "21.5" "33.799999999999997"
[87] "35" "31.100000000000001"
[89] "29.300000000000001" "40.100000000000001"
[91] "35.399999999999999" ":"
[93] "27.699999999999999" "70.400000000000006"
[95] "43.899999999999999" ":"
[97] "27" "38"
[99] "31.5" "47.600000000000001"
[101] "38.299999999999997" "16.5"
[103] "37.5" "56.100000000000001"
[105] "77.5" "75.700000000000003"
[107] "35.100000000000001" "28.600000000000001"
[109] "21" "16.600000000000001"
[111] ":" ":"
[113] ":" "18.899999999999999"
[115] "13" "21.100000000000001"
[117] "16" "21.100000000000001"
[119] "22.5" ":"
[121] "114.59999999999999" "83"
[123] ":" ":"
[125] ":" ":"
[127] "107" "132.5"
[129] ":" ":"
[131] ":" ":"
[133] ":" ":"
[135] ":" NA
[137] NA "not available"
[139] NA "Late foetal mortality rate"
[141] "Rate" NA
[143] "1960" ":"
[145] ":" ":"
[147] ":" ":"
[149] "15.300000000000001" "12.199999999999999"
[151] "9.8000000000000007" "12.4"
[153] "15.300000000000001" "15.5"
[155] "13" "21.899999999999999"
[157] "14.300000000000001" "27.300000000000001"
[159] ":" "17"
[161] "12.4" "24.5"
[163] ":" "10.699999999999999"
[165] "8.9000000000000004" "16.100000000000001"
[167] "13.199999999999999" ":"
[169] "14.9" "15"
[171] "12.5" "26.5"
[173] "15.9" "14.199999999999999"
[175] "10.9" "15.1"
[177] "13.699999999999999" ":"
[179] ":" ":"
[181] "12.4" "12.699999999999999"
[183] "10.4" "13.9"
[185] "11.4" "20.100000000000001"
[187] ":" "9.8000000000000007"
[189] ":" ":"
[191] ":" ":"
[193] ":" "8.6999999999999993"
[195] ":" ":"
[197] ":" ":"
[199] ":" ":"
[201] ":" ":"
[203] NA NA
[205] "not available" NA
[207] "Neonatal mortality rate" "Rate"
[209] NA "1960"
[211] ":" ":"
[213] ":" ":"
[215] ":" "20.5"
[217] "19.399999999999999" "13.1"
[219] "16.100000000000001" "23.899999999999999"
[221] "23.199999999999999" ":"
[223] "20.399999999999999" "19.5"
[225] "20.199999999999999" ":"
[227] "17.699999999999999" "35.100000000000001"
[229] "23.899999999999999" ":"
[231] "10.9" "13.4"
[233] "19.100000000000001" "27"
[235] ":" "13.5"
[237] "24.600000000000001" ":"
[239] "27.899999999999999" ":"
[241] "20.399999999999999" "14.1"
[243] "14.4" "13.4"
[245] ":" ":"
[247] ":" ":"
[249] "9.1999999999999993" ":"
[251] "11.699999999999999" "16.100000000000001"
[253] "16" ":"
[255] "41.399999999999999" ":"
[257] ":" ":"
[259] ":" ":"
[261] "32.799999999999997" ":"
[263] ":" ":"
[265] ":" ":"
[267] ":" ":"
[269] ":" NA
[271] NA "not available"
[273] NA "Perinatal mortality rate"
[275] "Rate" NA
[277] "1960" ":"
[279] ":" ":"
[281] ":" ":"
[283] "32.100000000000001" "23"
[285] "20.5" "26.199999999999999"
[287] "35.799999999999997" "34.899999999999999"
[289] "22.5" "37.700000000000003"
[291] "26.399999999999999" "42.799999999999997"
[293] ":" "31.399999999999999"
[295] "33.100000000000001" "41.899999999999999"
[297] ":" ":"
[299] "16.100000000000001" "32.200000000000003"
[301] "35" ":"
[303] "26.600000000000001" "34.899999999999999"
[305] ":" "41.100000000000001"
[307] ":" "29.600000000000001"
[309] "21" "27.5"
[311] "25.399999999999999" ":"
[313] ":" ":"
[315] ":" "19.699999999999999"
[317] ":" "23.699999999999999"
[319] "25.600000000000001" "33.5"
[321] ":" "28.199999999999999"
[323] ":" ":"
[325] ":" ":"
[327] ":" "25.800000000000001"
[329] ":" ":"
[331] ":" ":"
[333] ":" ":"
[335] ":" ":"
[337] NA NA
[339] "not available" After exploring the data, you may realise that the name of the data is always in the second column. Our data starts where find we the “Infant mortality rate”. The first row of the table where we find “GEO/TIME” in the first column
data_start_index <- pull(infant_mortality_df, 2) %>%
purrr::detect_index(~ . == "Infant mortality rate" & !is.na(.))
data_start_index[1] 73And ends at the next empty cell (not “:”)
infant_mortality_df %>%
slice(start_index:end_index)# A tibble: 61 × 62
Infant mortality rate…¹ ...2 ...3 ...4 ...5 ...6 ...7 ...8 ...9 ...10
<chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 GEO/TIME 1960 1961 1962 1963 1964 1965 1966 1967 1968
2 European Union - 27 co… : 38.2… 36.3… 34.2… 31.8… 30 29.1… 28.6… 29.1…
3 European Union - 28 co… : 36.2… 34.6… 32.7… 30.5 28.6… 28 27.5 27.8…
4 European Union - 27 co… : 36 34.3… 32.5 30.3… 28.5 27.8… 27.3… 27.8…
5 Euro area - 19 countri… : 34.7… 33.2… 31.8… 29.6… 28.3… 27.6… 26.1… 25.6…
6 Euro area - 18 countri… : 34.7… 33.2… 31.8… 29.6… 28.3… 27.6… 26.1… 25.6…
7 Belgium 31.3… 28.1… 27.5 27.1… 25.3… 23.6… 24.6… 22.8… 21.6…
8 Bulgaria 45.1… 37.7… 37.2… 35.7… 32.8… 30.8… 32.2… 33.1… 28.3…
9 Czechia 20 19.3… 21.1… 19.6… 19.1… 23.6… 21.8… 21.5 21.6…
10 Denmark 21.5 21.8… 20.1… 19.1… 18.6… 18.6… 16.8… 15.8… 16.3…
# ℹ 51 more rows
# ℹ abbreviated name: ¹`Infant mortality rates [demo_minfind]`
# ℹ 52 more variables: ...11 <chr>, ...12 <chr>, ...13 <chr>, ...14 <chr>,
# ...15 <chr>, ...16 <chr>, ...17 <chr>, ...18 <chr>, ...19 <chr>,
# ...20 <chr>, ...21 <chr>, ...22 <chr>, ...23 <chr>, ...24 <chr>,
# ...25 <chr>, ...26 <chr>, ...27 <chr>, ...28 <chr>, ...29 <chr>,
# ...30 <chr>, ...31 <chr>, ...32 <chr>, ...33 <chr>, ...34 <chr>, …If the colnames are not tidy or they are threated as observation, then use the janitor package.
infant_mortality_df %>%
slice(start_index:end_index) %>%
janitor::row_to_names(1)# A tibble: 60 × 62
`GEO/TIME` `1960` `1961` `1962` `1963` `1964` `1965` `1966` `1967` `1968`
<chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 European Unio… : 38.20… 36.39… 34.29… 31.89… 30 29.19… 28.69… 29.19…
2 European Unio… : 36.20… 34.60… 32.70… 30.5 28.69… 28 27.5 27.89…
3 European Unio… : 36 34.39… 32.5 30.30… 28.5 27.80… 27.39… 27.80…
4 Euro area - 1… : 34.79… 33.29… 31.80… 29.60… 28.30… 27.60… 26.19… 25.69…
5 Euro area - 1… : 34.79… 33.20… 31.80… 29.60… 28.30… 27.60… 26.19… 25.69…
6 Belgium 31.39… 28.10… 27.5 27.19… 25.30… 23.69… 24.69… 22.89… 21.69…
7 Bulgaria 45.10… 37.79… 37.29… 35.70… 32.89… 30.80… 32.20… 33.10… 28.30…
8 Czechia 20 19.30… 21.10… 19.69… 19.10… 23.69… 21.89… 21.5 21.60…
9 Denmark 21.5 21.80… 20.10… 19.10… 18.69… 18.69… 16.89… 15.80… 16.39…
10 Germany (unti… 33.79… 31.69… 29.30… 27 25.30… 23.89… 23.60… 22.89… 22.80…
# ℹ 50 more rows
# ℹ 52 more variables: `1969` <chr>, `1970` <chr>, `1971` <chr>, `1972` <chr>,
# `1973` <chr>, `1974` <chr>, `1975` <chr>, `1976` <chr>, `1977` <chr>,
# `1978` <chr>, `1979` <chr>, `1980` <chr>, `1981` <chr>, `1982` <chr>,
# `1983` <chr>, `1984` <chr>, `1985` <chr>, `1986` <chr>, `1987` <chr>,
# `1988` <chr>, `1989` <chr>, `1990` <chr>, `1991` <chr>, `1992` <chr>,
# `1993` <chr>, `1994` <chr>, `1995` <chr>, `1996` <chr>, `1997` <chr>, …Next issue: All columns are in character format
infant_mortality_df %>%
slice(start_index:end_index) %>%
janitor::row_to_names(1) %>%
* mutate_at(- 1, as.numeric) # all except the 1st col# A tibble: 60 × 62
`GEO/TIME` `1960` `1961` `1962` `1963` `1964` `1965` `1966` `1967` `1968`
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 European Unio… NA 38.2 36.4 34.3 31.9 30 29.2 28.7 29.2
2 European Unio… NA 36.2 34.6 32.7 30.5 28.7 28 27.5 27.9
3 European Unio… NA 36 34.4 32.5 30.3 28.5 27.8 27.4 27.8
4 Euro area - 1… NA 34.8 33.3 31.8 29.6 28.3 27.6 26.2 25.7
5 Euro area - 1… NA 34.8 33.2 31.8 29.6 28.3 27.6 26.2 25.7
6 Belgium 31.4 28.1 27.5 27.2 25.3 23.7 24.7 22.9 21.7
7 Bulgaria 45.1 37.8 37.3 35.7 32.9 30.8 32.2 33.1 28.3
8 Czechia 20 19.3 21.1 19.7 19.1 23.7 21.9 21.5 21.6
9 Denmark 21.5 21.8 20.1 19.1 18.7 18.7 16.9 15.8 16.4
10 Germany (unti… 33.8 31.7 29.3 27 25.3 23.9 23.6 22.9 22.8
# ℹ 50 more rows
# ℹ 52 more variables: `1969` <dbl>, `1970` <dbl>, `1971` <dbl>, `1972` <dbl>,
# `1973` <dbl>, `1974` <dbl>, `1975` <dbl>, `1976` <dbl>, `1977` <dbl>,
# `1978` <dbl>, `1979` <dbl>, `1980` <dbl>, `1981` <dbl>, `1982` <dbl>,
# `1983` <dbl>, `1984` <dbl>, `1985` <dbl>, `1986` <dbl>, `1987` <dbl>,
# `1988` <dbl>, `1989` <dbl>, `1990` <dbl>, `1991` <dbl>, `1992` <dbl>,
# `1993` <dbl>, `1994` <dbl>, `1995` <dbl>, `1996` <dbl>, `1997` <dbl>, …And now lets transform it into long format.
infant_mortality_df %>%
slice(start_index:end_index) %>%
janitor::row_to_names(1) %>%
mutate_at(- 1, as.numeric) %>%
* pivot_longer(- 1, names_to = "TIME",
values_to = "infant_mortality")# A tibble: 3,660 × 3
`GEO/TIME` TIME infant_mortality
<chr> <chr> <dbl>
1 European Union - 27 countries (from 2020) 1960 NA
2 European Union - 27 countries (from 2020) 1961 38.2
3 European Union - 27 countries (from 2020) 1962 36.4
4 European Union - 27 countries (from 2020) 1963 34.3
5 European Union - 27 countries (from 2020) 1964 31.9
6 European Union - 27 countries (from 2020) 1965 30
7 European Union - 27 countries (from 2020) 1966 29.2
8 European Union - 27 countries (from 2020) 1967 28.7
9 European Union - 27 countries (from 2020) 1968 29.2
10 European Union - 27 countries (from 2020) 1969 28
# ℹ 3,650 more rowsLets filter out aggregates and irrelevant.
infant_mortality_df %>%
slice(start_index:end_index) %>%
janitor::row_to_names(1) %>%
mutate_at(- 1, as.numeric) %>%
pivot_longer(- 1, names_to = "TIME",
values_to = "infant_mortality") %>%
rename(geo = `GEO/TIME`) %>%
mutate(
geo = countrycode(geo, "country.name", "iso3c"),
) %>%
filter(!is.na(geo))This code filters out the rows where geo is in:
Enough?
count() Quickly count the unique values of one or more variables
infant_mortality_df %>%
slice(start_index:end_index) %>%
janitor::row_to_names(1) %>%
mutate_at(- 1, as.numeric) %>%
pivot_longer(- 1, names_to = "TIME",
values_to = "infant_mortality") %>%
rename(geo = `GEO/TIME`) %>%
mutate(
geo = countrycode(geo, "country.name", "iso3c"),
) %>%
filter(!is.na(geo)) %>%
count(geo, sort = TRUE)# A tibble: 47 × 2
geo n
<chr> <int>
1 DEU 122
2 FRA 122
3 ALB 61
4 AND 61
5 ARM 61
6 AUT 61
7 AZE 61
8 BEL 61
9 BGR 61
10 BIH 61
# ℹ 37 more rowsRemove the irrelevant duplication & convert TIME to numeric
infant_mortality_df %>%
slice(start_index:end_index) %>%
janitor::row_to_names(1) %>%
mutate_at(- 1, as.numeric) %>%
pivot_longer(- 1, names_to = "TIME",
values_to = "infant_mortality") %>%
rename(geo = `GEO/TIME`) %>%
filter(
geo != "France (metropolitan)" &
geo != "Germany (until 1990 former territory of the FRG)"
) %>%
mutate(
geo = countrycode(geo, "country.name", "iso3c"),
TIME = as.numeric(TIME)
) %>%
filter(!is.na(geo)) # A tibble: 2,867 × 3
geo TIME infant_mortality
<chr> <dbl> <dbl>
1 BEL 1960 31.4
2 BEL 1961 28.1
3 BEL 1962 27.5
4 BEL 1963 27.2
5 BEL 1964 25.3
6 BEL 1965 23.7
7 BEL 1966 24.7
8 BEL 1967 22.9
9 BEL 1968 21.7
10 BEL 1969 21.2
# ℹ 2,857 more rowsAssign it with the same name (re-write the table)
infant_mortality_df <- infant_mortality_df %>%
slice(start_index:end_index) %>%
janitor::row_to_names(1) %>%
mutate_at(- 1, as.numeric) %>%
pivot_longer(- 1, names_to = "TIME",
values_to = "infant_mortality") %>%
rename(geo = `GEO/TIME`) %>%
filter(
geo != "France (metropolitan)" &
geo != "Germany (until 1990 former territory of the FRG)"
) %>%
mutate(
geo = countrycode(geo, "country.name", "iso3c"),
TIME = as.numeric(TIME)
) %>%
filter(!is.na(geo)) Let’s transform fertility_df to a similar format
fertility_df <- read_csv("https://stats.oecd.org/sdmx-json/data/DP_LIVE/.FERTILITY.../OECD?contentType=csv&detail=code&separator=comma&csv-lang=en")fertility_df <- fertility_df %>%
select(
geo = LOCATION, TIME, fertility = Value
)Now we need to link the two tables.
df <- full_join(x = fertility_df, y = infant_mortality_df)
df# A tibble: 4,205 × 4
geo TIME fertility infant_mortality
<chr> <dbl> <dbl> <dbl>
1 AUS 1960 3.45 NA
2 AUS 1961 3.55 NA
3 AUS 1962 3.43 NA
4 AUS 1963 3.34 NA
5 AUS 1964 3.15 NA
6 AUS 1965 2.97 NA
7 AUS 1966 2.89 NA
8 AUS 1967 2.85 NA
9 AUS 1968 2.89 NA
10 AUS 1969 2.89 NA
# ℹ 4,195 more rowsAnd now… Let’s see what we have here. Last time we saw that we can generate summary statistics with skimr function.
skimr::skim(df)| Name | df |
| Number of rows | 4205 |
| Number of columns | 4 |
| _______________________ | |
| Column type frequency: | |
| character | 1 |
| numeric | 3 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| geo | 0 | 1 | 2 | 4 | 0 | 68 | 0 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| TIME | 0 | 1.00 | 1990.42 | 17.85 | 1960.00 | 1975.00 | 1990.00 | 2006.00 | 2022.00 | ▇▇▇▇▇ |
| fertility | 911 | 0.78 | 2.34 | 1.20 | 0.81 | 1.59 | 1.94 | 2.59 | 7.67 | ▇▃▁▁▁ |
| infant_mortality | 1890 | 0.55 | 14.33 | 15.03 | 0.00 | 4.90 | 9.70 | 17.90 | 137.60 | ▇▁▁▁▁ |
We can also generate some pots and statistics easily with the powerful radiant package.
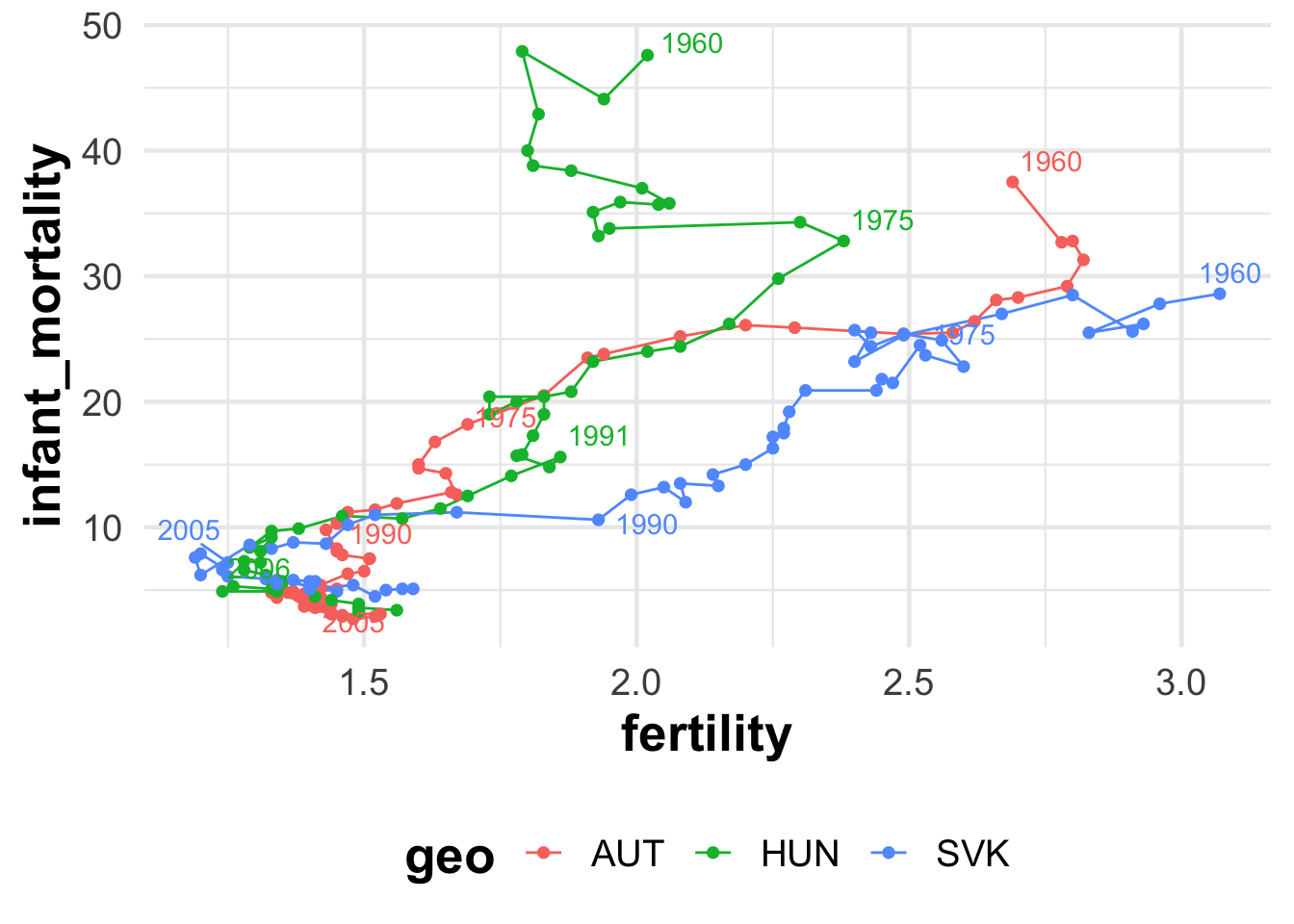
radiant::radiant() # start a shiny app on your machineBut life is never so easy. Let’s suppose we want to visualise only a few countries to tell a story.
beveridge_plot <- function(data, x, y, group, time, n_label = 5) {
data |>
arrange({{ time }}) |>
ggplot() +
aes({{ x }}, {{ y }}, color = {{ group }}) +
geom_path() +
geom_point(size = 2, shape = 16) +
ggrepel::geom_text_repel(data = ~ group_by(., {{ group }}) |>
slice(unique(floor(seq(from = 1, to = n(),
length.out = n_label)))),
aes(label = {{ time }}),
show.legend = FALSE
)
}